Imports:
Python:
import jax
import jax.numpy as jnp
import numpyro
from numpyro import distributions as dist, inferimport pandas as pd
import matplotlib.pyplot as plt
import arviz as az
import seaborn as snsData:
# BDA3 table 5.1 data: rat tumor
tumors = jnp.array([
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1,
1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 1, 5, 2,
5, 3, 2, 7, 7, 3, 3, 2, 9, 10, 4, 4, 4, 4, 4, 4, 4,
10, 4, 4, 4, 5, 11, 12, 5, 5, 6, 5, 6, 6, 6, 6, 16, 15,
15, 9, 4
])
pop = jnp.array([
20, 20, 20, 20, 20, 20, 20, 19, 19, 19, 19, 18, 18, 17, 20, 20, 20,
20, 19, 19, 18, 18, 25, 24, 23, 20, 20, 20, 20, 20, 20, 10, 49, 19,
46, 27, 17, 49, 47, 20, 20, 13, 48, 50, 20, 20, 20, 20, 20, 20, 20,
48, 19, 19, 19, 22, 46, 49, 20, 20, 23, 19, 22, 20, 20, 20, 52, 46,
47, 24, 14
])Model:
The natural model here is a binomial model, with a beta prior on the probability of a tumours (), so that we can exploit conjugacy if we want to. That is, we have the model:
In this case, we have prior information in the form of previous experiments which use the same variant of rat. In these experiments, we expect the latent probability of a tumour to vary according to the experiment – it is probably a function of environment, diet, and so on.
We denote these previous experiments as , such that our current trial is , corresponding to the scenario as originally outlined.
We can incorporate these other experiments into our model by estimating a unique for each experiment, with the values of drawn from a common prior over values. This extends the above model to:
We need to specify a hyperprior over and –- BDA3 contains a long discussion on this issue. We choose to parametrize the mean and variance of the distribution, transforming to and after sampling.
Our NumPyro model here is:
def rat_mod_full(y=None, n=None):
# Hyperpriors
mu = numpyro.sample("mu", dist.Beta(2, 2))
tau = numpyro.sample("tau", dist.Gamma(2, 0.5))
# Transformation to alpha, beta
alpha = mu * tau
beta = (1-mu) * tau
# Prior on the probability of a tumour
with numpyro.plate("theta_plate", len(pop)):
theta = numpyro.sample("theta", dist.Beta(alpha, beta))
# Likelihood
with numpyro.plate("obs", len(pop)):
numpyro.sample("y", dist.Binomial(n, theta), obs = y)Inference:
We use NUTS to sample from the posterior:
sampler = infer.MCMC(
infer.NUTS(rat_mod_full),
num_warmup=500,
num_samples=1000,
num_chains=1,
)
sampler.run(
jax.random.PRNGKey(1),
y=jnp.array(tumors),
n=pop
)
samples = sampler.get_samples()Diagnostics:
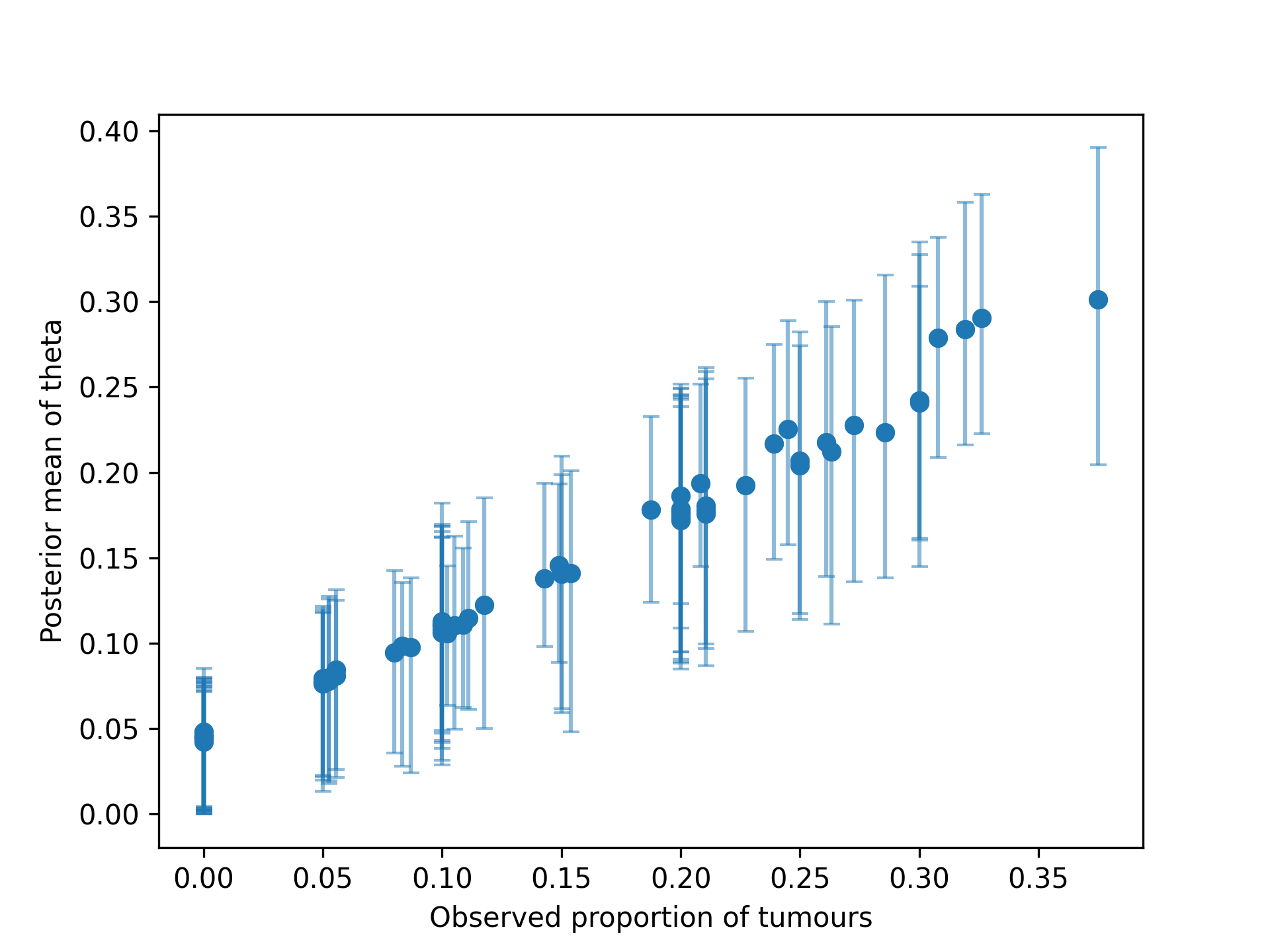
We can reproduce Figure 5.4 from BDA3: